Projects in Biomaterial Science
Prediction of fibrinogen adsorption for biodegradable polymers:
Integration of molecular dynamics and surrogate modeling
Integration of molecular dynamics and surrogate modeling
(with D. KNIGHT - Department of Mechanical and Aerospace Engineering, Rutgers, The State University of New Jersey, NJ USA; V. KHOLODOVYCH & W. J. WELSH - Department of Pharmacology, University of Medicine and Dentistry of New Jersey, NJ USA and J. KOHN$ - New Jersey Center for Biomaterials, Rutgers, The State University of New Jersey, NJ USA)
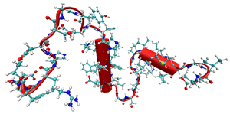
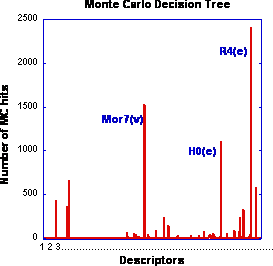
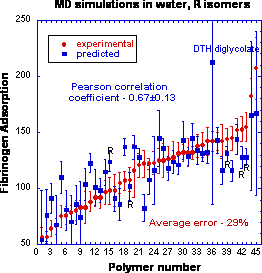
This work is a part of a series of publications devoted to the development of surrogate (semi-empirical) models for the prediction of fibrinogen adsorption onto polymer surfaces. Since fibrinogen is one of the key proteins involved in platelet activation and the formation of thrombosis, the modeling of fibrinogen adsorption on the surface of blood-contacting medical devices is of high theoretical and practical significance. We report here, for the first time, on the incorporation of three-dimensional structures of polymers obtained from atomistic simulations into conventional mesoscopic-scale calculations. Low energy conformations derived from molecular dynamics simulations for 45 representatives of a combinatorial library of polyarylates were used in an improved modeling procedure (referred to as ‘‘3D surrogate model’’) instead of simplistic two-dimensional representations of polymer structures, which were used in several previous models (collectively referred to as ‘‘2D surrogate models’’). In the framework of this 3D model we created 12 model sets of polymers to account for their chirality, conformational diversity and the structural influence of a solvent. For each polymer set, three-dimensional molecular descriptors were generated and then ranked with respect to the experimental fibrinogen adsorption data by means of a Monte Carlo decision tree. The most significant descriptors identified by decision tree and the experimental dataset were utilized to predict fibrinogen adsorption using an artificial neural network (ANN). The best prediction achieved by the 3D surrogate model demonstrated a noticeable improvement in the predictive quality as compared to the previously used 2D model (as evidenced by the increase in the average Pearson correlation coefficient from 0.54 ± 0.12 to 0.67 ± 0.13). The predictive quality of the 3D surrogate model compares favorably with the best results previously reported for extended 2D model that combines an ANN with partial least squares (PLS) regression and principal component (PC) analysis. The significance of the newly developed 3D model is that it allows high accuracy prediction of fibrinogen adsorption without the need for experimentally-derived descriptors and it has better predictive quality than the original 2D surrogate model due to utilization of realistic polymer representations.
 |
 |
Logical analysis of data in structure-activity investigation
of polymeric gene delivery
of polymeric gene delivery
(with T. O. BONATES - Rutgers University Center for Operations Research, Rutgers, The State University of New Jersey, NJ USA; V. KHOLODOVYCH - Department of Pharmacology, University of Medicine and Dentistry of New Jersey, NJ USA; R. LANGER - Department of Chemical Engineering, Massachusetts Institute of Technology, MA USA and J. KOHN$ - New Jersey Center for Biomaterials, Rutgers, The State University of New Jersey, NJ USA)
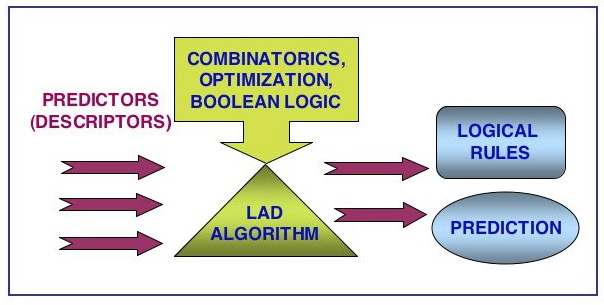
To date semi-empirical or surrogate modeling demonstrated an obvious success in prediction of various kinds of biological response. In the present work the potential of semi-empirical modeling in identification of the most promising candidates for polymeric gene delivery was evaluated on the dataset published by Langer and co-workers. For the first time, a correlation between chemical structures of poly(b-amino esters) and their efficiency in transfecting DNA was established using the novel technique of logical analysis of data (LAD). It became possible due to utilization of the new LAD regression algorithm that allows capturing originally “hidden” correlations between input variables. Indeed, the employed LAD methodology confirmed its superiority with respect to such well-established methods as Polynomial and Artificial Neural Networks and Support Vector Machines. The most successful regression function was associated with Pearson correlation coefficient of 0.77 and mean absolute error of 3.83. The corresponding values of predicted transfection efficiency were in a very good agreement with experimentally measured values for this type of bioresponce. Two modeling approaches, particularly linear combination and explicit representation models were introduced and compared in the framework of the present study. A set of molecular descriptors was selected and tested on its ability to identify important structural features and physicochemical properties associated with the most efficient in terms of DNA delivery poly(b-amino esters). Finally, it was shown that detailed analysis of the rules provided by LAD algorithm could serve as an excellent guide to a polymer chemist in the design of new biomaterials.
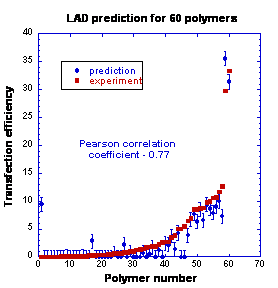 |
 |
Virtual combinatorial library of polymethacrylates
(with D. KNIGHT - Department of Mechanical and Aerospace Engineering, Rutgers, The State University of New Jersey, NJ USA and J. KOHN$ - New Jersey Center for Biomaterials, Rutgers, The State University of New Jersey, NJ USA)
A large (about 40000 compounds) virtual combinatorial library of polymethacrylates was, for the first time, designed for computer-aided prediction of biological and material properties and focused polymer synthesis. To introduce structural diversity into our combinatorial design, 33 building blocks (side chains) from a family of methacrylates were chosen as candidates for “computational combinatorial synthesis”. A length of the common structural skeleton (polymer backbone) was built to accommodate 12 repeat units to encompass variations in both co- and terpolymer compositions. The Molecular Operating Environment (MOE) modeling package from Chemical Computing Group Inc. was used to generate a virtual library of about 40000 polymers, where the building blocks were randomly combined to reproduce 50/50, 25/75, 75/25, and 33/33/33 percent compositions for co- and terpolymers, respectively. The entire combinatorial virtual library included all possible permutations and comprised three sub-libraries: (a) homopolymers of 33 polymers, (b) copolymers of 3267 (33x33) polymers and (c) terpolymers of 35937 (33x33x33) structures. Schematic representation of this virtual library is shown in figure below. The binomial coefficient was used to select unique polymers by reducing the total number of copolymer structures from 1089 to 528 for each of the 50/50, 25/75 and 75/25 percent compositions (total 1584 compounds). Similarly, the 35937 terpolymers were reduced to 5456 unique structures and the dataset of 330 terpolymers was employed in subsequent QSAR modeling. Fully automated energy minimization was performed using the MMFF force field to ensure the correct geometry of all polymers. The exceptional large size, structural diversity, and explicit representation of polymer compositions make this virtual library of polymethacrylates an ideal test case for computer-aided prediction of important characteristics of biodegradable polymers.
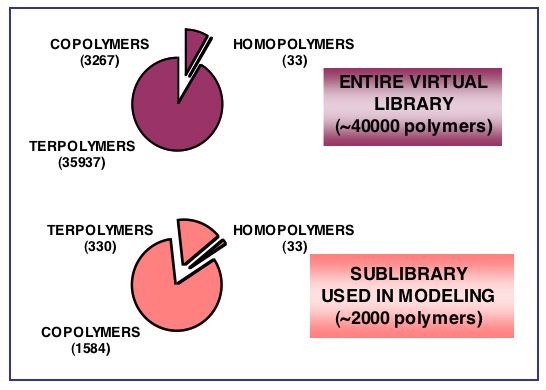
QSAR modeling in prediction of biological response for large combinatorial library
of biodegradable polymers (project in progress)
(with V. KHOLODOVYCH & W. J. WELSH - Department of Pharmacology, University of Medicine and Dentistry of New Jersey, NJ USA; D. KNIGHT - Department of Mechanical and Aerospace Engineering, Rutgers, The State University of New Jersey, NJ USA; and J. KOHN$ - New Jersey Center for Biomaterials, Rutgers, The State University of New Jersey, NJ USA)
A subset of 79 polymers taken from a representative sub-library of 2000 polymethacrylates was employed to build initial QSAR-based models, which were then deployed to predict cell attachment, cell growth, and fibrinogen adsorption on polymer surfaces for these 2000 polymethacrylates. The Polynomial Neural Network – a powerful machine-learning algorithm has been used to this end. The agreement between predicted and experimentally measured property values for the 50 polymethacrylate copolymers within this virtual polymer space encourages further pursuit of polymethacrylate-based biomaterials, and justifies more extensive deployment of computational models derived from larger experimental data sets for the rational design of biorelevant polymers endowed with targeted performance properties.
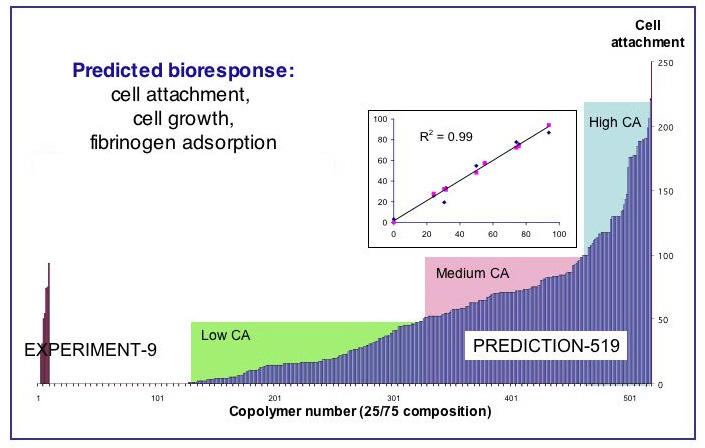
Experimental and computational study of controlled release of a hydrophobic peptide from matrices of biodegradable polymers
(with I.J. KHAN - Department of Biochemistry, Center for Advanced Biotechnology and Medicine, University of Medicine and Dentistry of New Jersey, NJ USA; L.M. VALENZUELA - Department of Chemical and Bioprocess Engineering, Pontificia Universidad Católica de Chile, Santiago, Chile; Y.V. LISNYAK - Department of Molecular Modeling, Institute of Microbiology and Immunology, Kharkov, Ukraine; and J. KOHN$ - New Jersey Center for Biomaterials, Rutgers, The State University of New Jersey, NJ USA)
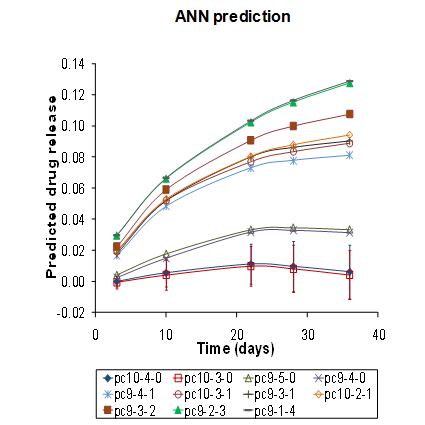
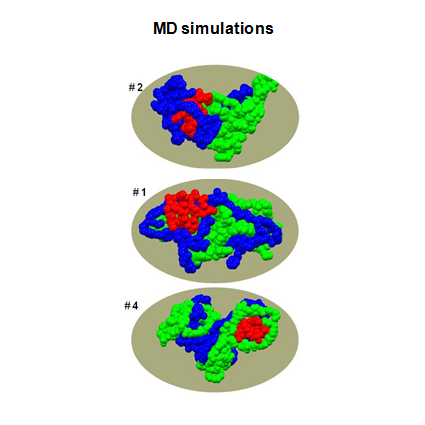
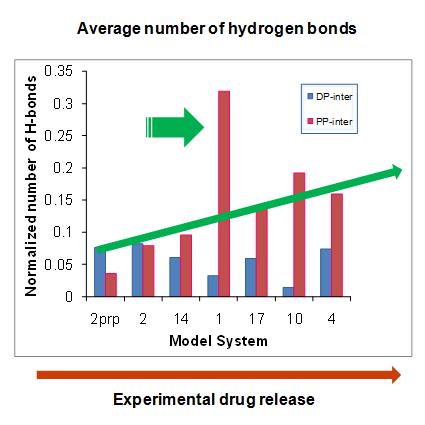
The objective of this work was two-fold: (1) to identify optimal compositions of tyrosine-derived poly(DTE-co-y% DT-co-z% PEG1K carbonate)s for controlled delivery of voclosporin, a potent drug candidate to treat ocular diseases and (2) to establish structure-function relationship between key molecular characteristics of biodegradable polymer matrices and drug release kinetics. For the first time, the experimental study of polymeric drug release was accompanied by hierarchical sequence of three computational methods. First, response surface methodology was used to select optimal polymer compositions for subsequent neural network modeling. Second, the accurate neural network models were built to predict drug release profiles for 15 polymers located outside of the initial design space. Finally, molecular dynamics simulations were carried out to study thermodynamic properties and hydrogen-bonding patterns of model drug-polymer complexes and to elucidate a role of specific interactions in drug release mechanism. This research presents further development of methodological approaches to meet the challenges in the design of polymeric drug delivery systems.
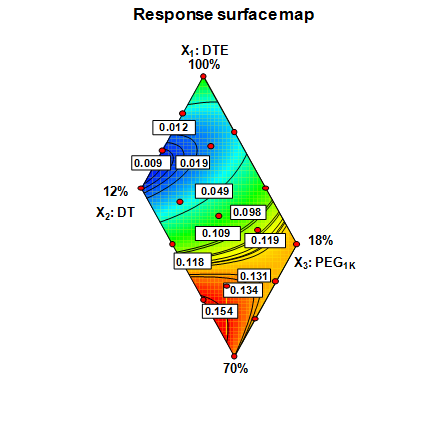 |
 |
 |
 |